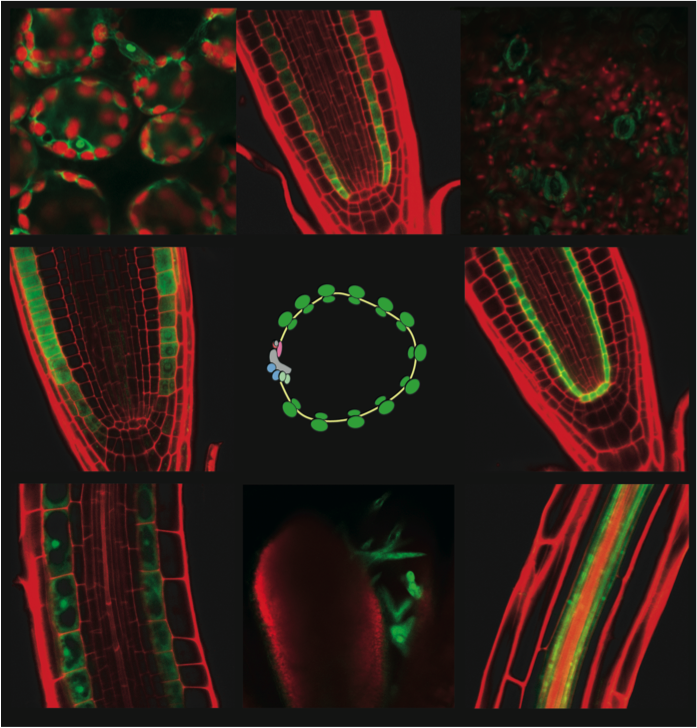
Aim 1: Establish INTACT and TRAP tools in tomato, rice and Medicago for comparison of the epigenome, transcriptome, and translatome for root and shoot meristem tissues.
We will produce genotypes for INTACT and TRAP targeted to actively dividing cells and the functional shoot and root apical meristems. With these we will compare the growth apices of different species using multiple genome scale readouts including the nuclear epigenome (H3K27me3 and H3K4me3), transcriptome, and translatome.
Progress: The binary backbone vectors for tomato, Medicago and rice have been generated. Through transient assays we have determined that species-specific components need to be incorporated including codon-optimized biotin ligase, nuclear envelope tags and constitutive promoters. Constructs are being introduced into their respective species at the UC Davis and UC Riverside Transformation Facilities.

